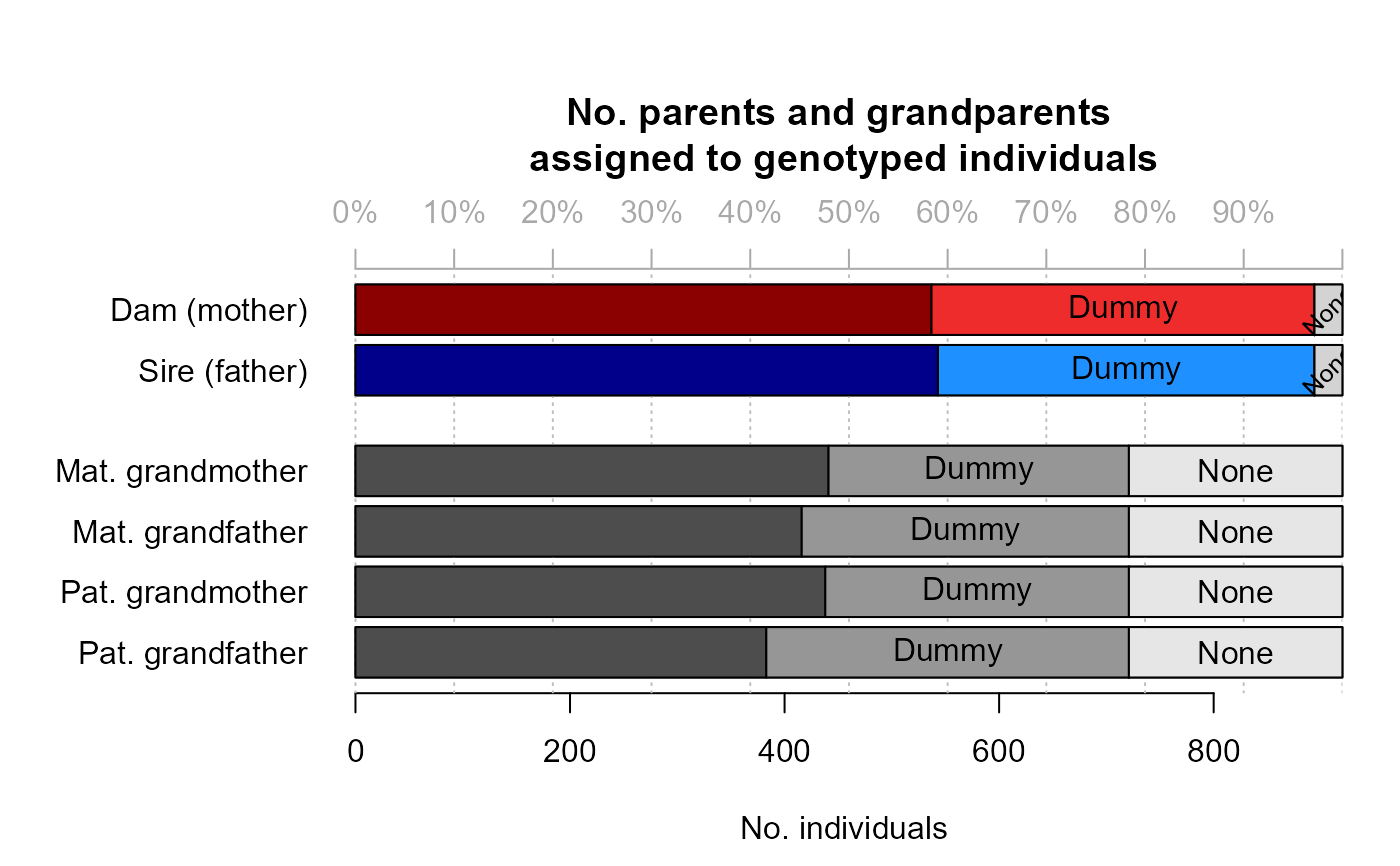
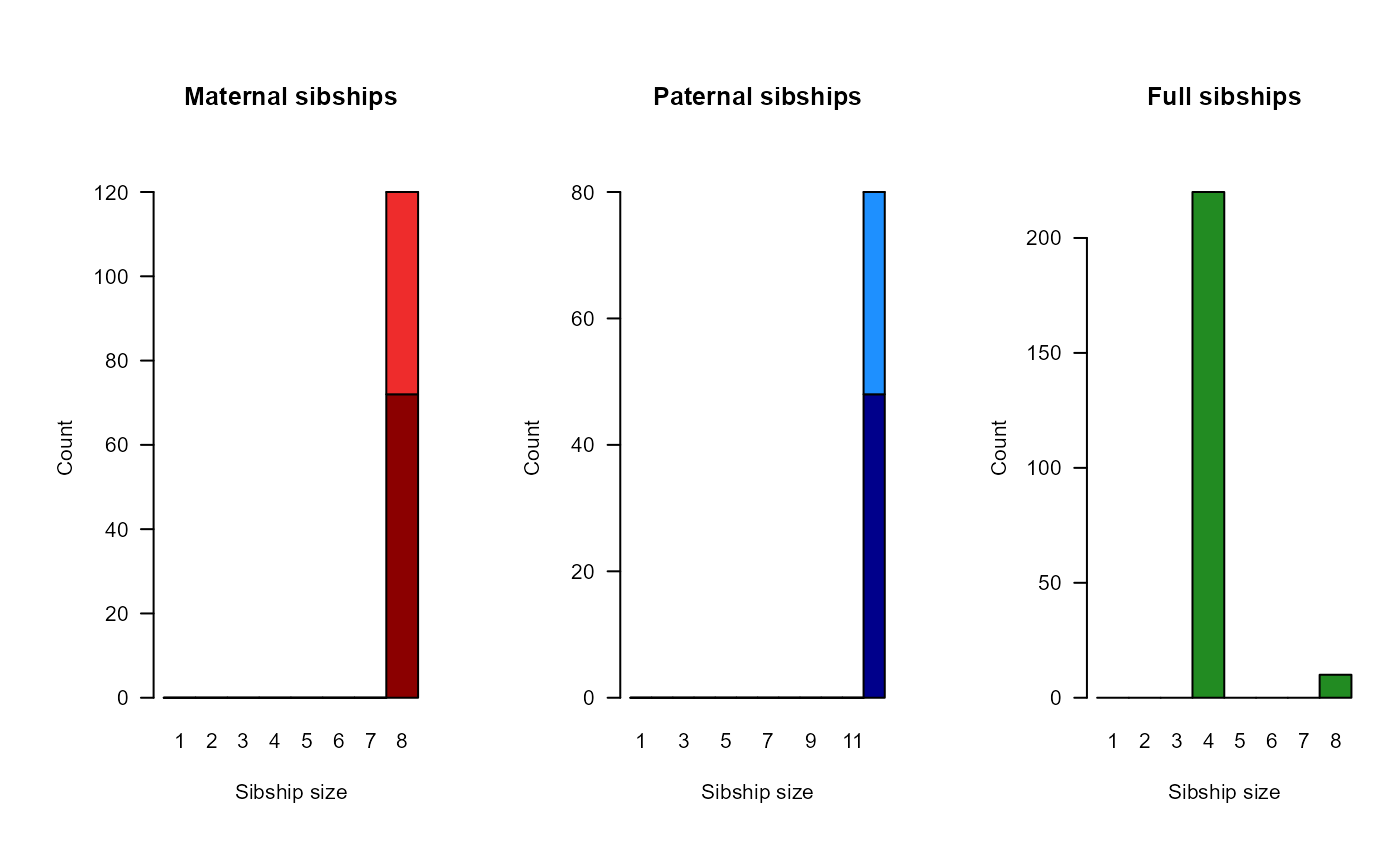
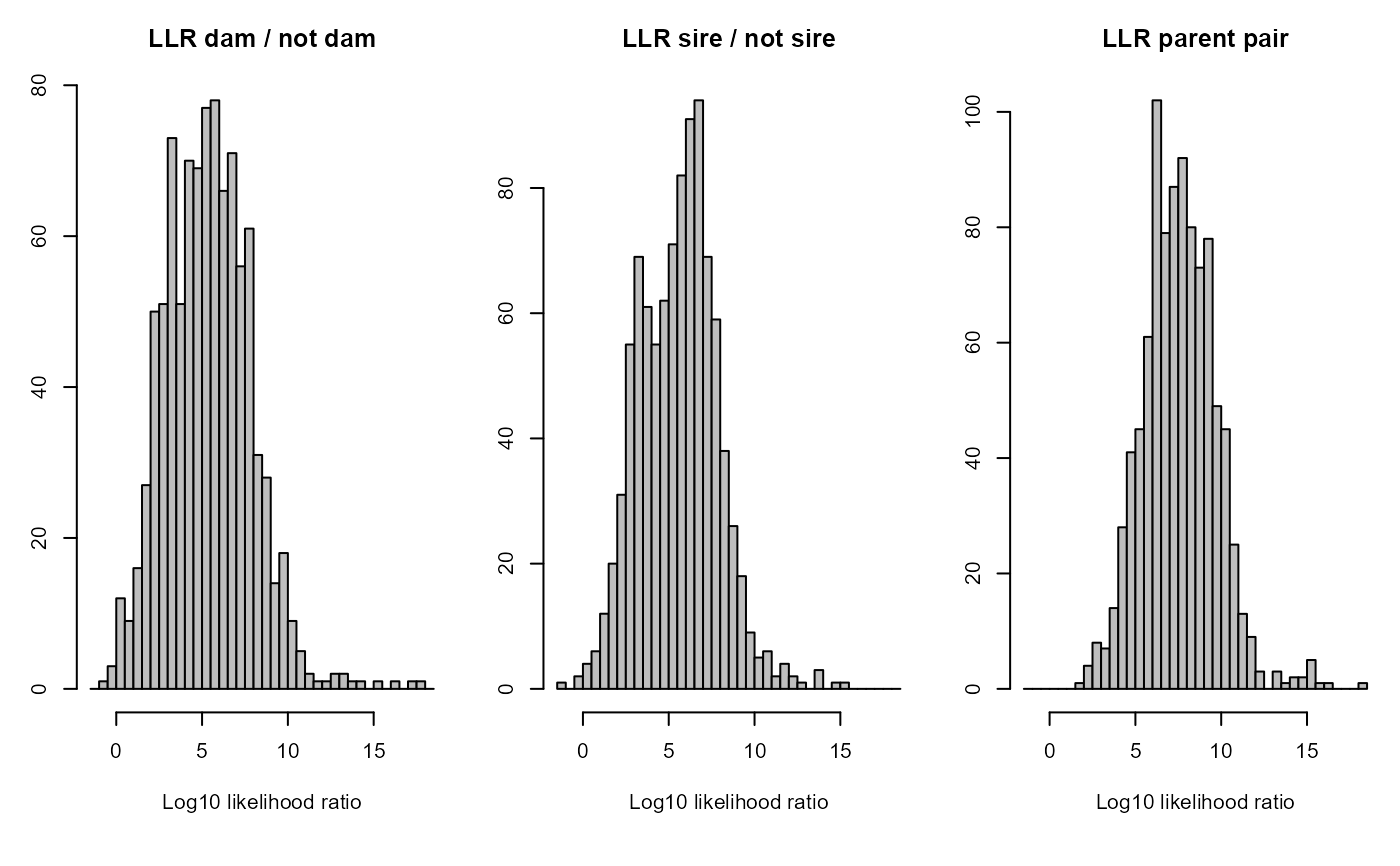
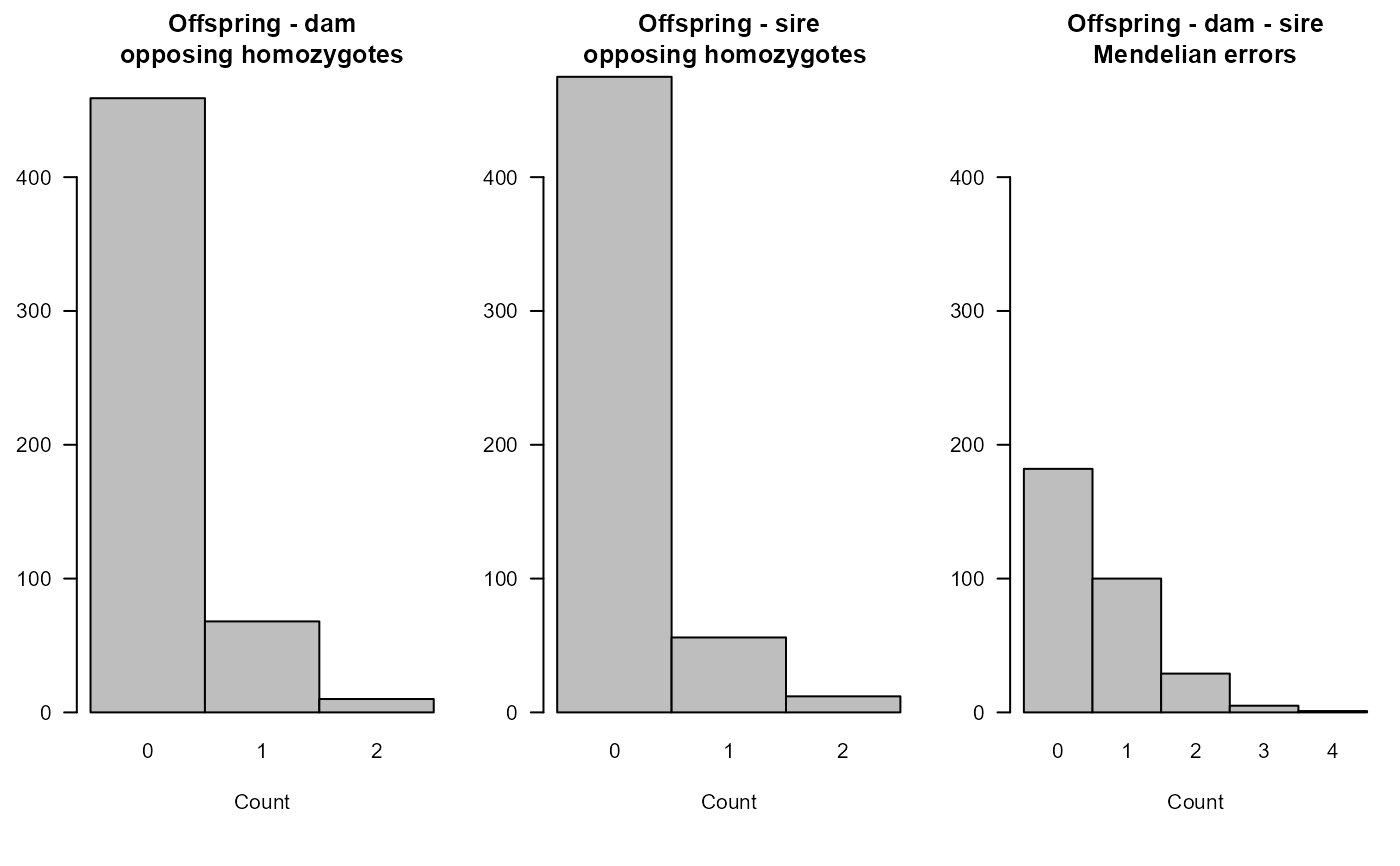Example output from pedigree inference: 'HSg5'
SeqOUT_HSg5.RdUsage
data(SeqOUT_HSg5)Format
a list, see sequoia
Author
Jisca Huisman, jisca.huisman@gmail.com
Examples
if (FALSE) { # \dontrun{
# this output was created as follows:
Geno <- SimGeno(Ped = Ped_HSg5, nSnp = 200)
SeqOUT_HSg5 <- sequoia(GenoM = Geno, LifeHistData = LH_HSg5, Module = "ped",
Err = 0.005)
} # }
# some ways to inspect the output; see vignette for more info:
names(SeqOUT_HSg5)
#> [1] "Specs" "ErrM" "args.AP" "Snps-LowCallRate"
#> [5] "AgePriors" "LifeHist" "DupLifeHistID" "NoLH"
#> [9] "PedigreePar" "TotLikPar" "LifeHistPar" "Pedigree"
#> [13] "DummyIDs" "TotLikSib" "AgePriorExtra" "LifeHistSib"
SeqOUT_HSg5$Specs
#> NumberIndivGenotyped NumberSnps GenotypingErrorRate MaxMismatchDUP
#> Specs 920 200 0.005 11
#> MaxMismatchOH MaxMismatchME Tfilter Tassign nAgeClasses MaxSibshipSize
#> Specs 5 8 -2 0.5 6 100
#> Module DummyPrefixFemale DummyPrefixMale Complexity Herm UseAge CalcLLR
#> Specs ped F M full no yes TRUE
#> ErrFlavour SequoiaVersion TimeStart TimeEnd
#> Specs version2.0 2.3.18 2022-12-17 13:26:47 2022-12-17 13:29:23
SummarySeq(SeqOUT_HSg5)